The Dynamic Adaptive Chemistry (DAC) method [1] simplify the chemistry using the matrix rAB defined by (DRGEP algorithm [2]) More...
Public Member Functions | |
| TypeName ("DAC") | |
| Runtime type information. More... | |
| DAC (const IOdictionary &dict, chemistryModel< ThermoType > &chemistry) | |
| Construct from components. More... | |
| virtual | ~DAC () |
| Destructor. More... | |
| virtual void | reduceMechanism (const scalar p, const scalar T, const scalarField &c, List< label > &ctos, DynamicList< label > &stoc, const label li) |
| Reduce the mechanism. More... | |
 Public Member Functions inherited from chemistryReductionMethod< ThermoType > Public Member Functions inherited from chemistryReductionMethod< ThermoType > | |
| TypeName ("chemistryReductionMethod") | |
| Runtime type information. More... | |
| declareRunTimeSelectionTable (autoPtr, chemistryReductionMethod, dictionary,(const IOdictionary &dict, chemistryModel< ThermoType > &chemistry),(dict, chemistry)) | |
| chemistryReductionMethod (chemistryModel< ThermoType > &chemistry) | |
| Construct from components. More... | |
| chemistryReductionMethod (const IOdictionary &dict, chemistryModel< ThermoType > &chemistry) | |
| Construct from components. More... | |
| virtual | ~chemistryReductionMethod () |
| Destructor. More... | |
| virtual bool | active () const |
| Return mechanism reduction active. More... | |
| label | nSpecie () |
| Return the number of species. More... | |
| label | nActiveSpecies () const |
| Return the number of active species. More... | |
| scalar | tolerance () const |
| Return the tolerance. More... | |
| const List< bool > & | activeSpecies () const |
| Return the active species. More... | |
| bool | reactionDisabled (const label i) const |
| Return whether or not a reaction is disabled. More... | |
| virtual void | update () |
| ... More... | |
Additional Inherited Members | |
 Static Public Member Functions inherited from chemistryReductionMethod< ThermoType > Static Public Member Functions inherited from chemistryReductionMethod< ThermoType > | |
| static autoPtr< chemistryReductionMethod< ThermoType > > | New (const IOdictionary &dict, chemistryModel< ThermoType > &chemistry) |
 Protected Member Functions inherited from chemistryReductionMethod< ThermoType > Protected Member Functions inherited from chemistryReductionMethod< ThermoType > | |
| void | initReduceMechanism () |
| Protected Member Functions. More... | |
| void | endReduceMechanism (List< label > &ctos, DynamicList< label > &stoc) |
| End reduction of the mechanism. More... | |
 Protected Attributes inherited from chemistryReductionMethod< ThermoType > Protected Attributes inherited from chemistryReductionMethod< ThermoType > | |
| const dictionary | coeffsDict_ |
| Dictionary that store the algorithm data. More... | |
| chemistryModel< ThermoType > & | chemistry_ |
| Reference to the chemistry model. More... | |
| const label | nSpecie_ |
| Total number of species. More... | |
| label | nActiveSpecies_ |
| Number of active species. More... | |
| Field< bool > | reactionsDisabled_ |
| List of disabled reactions (disabled = true) More... | |
| List< bool > | activeSpecies_ |
| List of active species (active = true) More... | |
The Dynamic Adaptive Chemistry (DAC) method [1] simplify the chemistry using the matrix rAB defined by (DRGEP algorithm [2])
|sum_i=1->Nr vAi wi dBi| rAB = ————————— , max(PA, CA)
PA = sum_i=1->Nr (max (0, vAi wi)) -> production of species A
CA = sum_i=1->Nr (max (0, -vAi wi)) -> consumption of species A
where i is the reaction index, Nr the number of reactions, vAi is the net stoichiometric coefficient of species A in the ith reaction (vAi = v''-v') , wi is the progress variable of reaction i and dBi equals 1 if reaction i involves B and O otherwise. rAB show the error introduced to the production rates of A when B and all the reactions including it are removed. It is computed as in [3] so that the algorithm is O(Nr).
DAC uses a initial set of species that represents the major parts of the combustion mechanism, i.e. H2/O2, fuel decomposition and CO2 production. Usually, it includes the fuel, HO2 and CO. Then it computes the dependence of these set to the other species. This is done by introducing R-value defined by
R_V0 (V) = max_SP(product(rij)),
where SP is the set of all possible paths leading from V0 to V and product(rij) is the chain product of the weights of the edges along the given path. The R-value for the initial set species is 1.
When the R-value of a species is larger than a user-defined tolerance then the species is included in the simplified mechanism. Otherwise, the species is removed along with all the reactions including it.
During this process, instead of looking over all species like described in [1], the algorithm implemented here creates dynamic list to retain the initialised edges only (see [3]).
References:
[1] Liang, L., Stevens, J. G., & Farrell, J. T. (2009).
A dynamic adaptive chemistry scheme for reactive flow computations.
Proceedings of the Combustion Institute, 32(1), 527-534.
[2] Pepiot-Desjardins, P., & Pitsch, H. (2008).
An efficient error-propagation-based reduction method for large
chemical kinetic mechanisms.
Combustion and Flame, 154(1), 67-81.
[3] Lu, T., & Law, C. K. (2006).
Linear time reduction of large kinetic mechanisms with directed
relation graph: n-Heptane and iso-octane.
Combustion and Flame, 144(1), 24-36.
| DAC | ( | const IOdictionary & | dict, |
| chemistryModel< ThermoType > & | chemistry | ||
| ) |
Construct from components.
Definition at line 32 of file DAC.C.
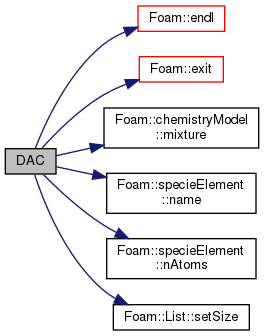
References Foam::endl(), Foam::exit(), Foam::FatalError, FatalErrorInFunction, forAll, forAllConstIter, Foam::Info, chemistryModel< ThermoType >::mixture(), specieElement::name(), specieElement::nAtoms(), nSpecie, and List< T >::setSize().
|
virtual |
Destructor.
Definition at line 228 of file DAC.C.
References DAC< ThermoType >::reduceMechanism().
| TypeName | ( | "DAC< ThermoType >" | ) |
Runtime type information.
|
virtual |
Reduce the mechanism.
Implements chemistryReductionMethod< ThermoType >.
Definition at line 236 of file DAC.C.
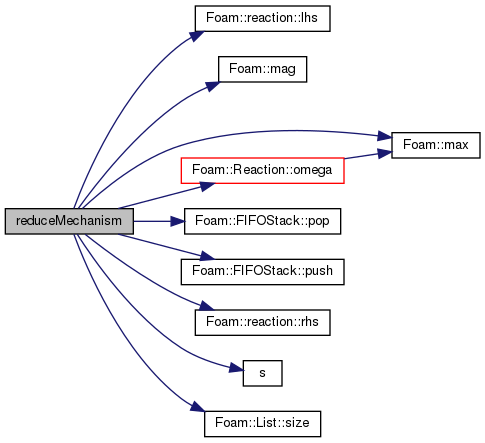
References Foam::constant::physicoChemical::c1, forAll, found, reaction::lhs(), Foam::mag(), Foam::max(), nSpecie, Reaction< ReactionThermo >::omega(), p, FIFOStack< T >::pop(), FIFOStack< T >::push(), R, reaction::rhs(), s(), List< T >::size(), and T.
Referenced by DAC< ThermoType >::~DAC().